GiniClust3: a fast and memory-efficient tool for rare cell type identification
GiniClust is a clustering method specifically designed for rare cell type detection. It uses the Gini index to identify genes that are associated with rare cell types without prior knowledge. This differs from traditional clustering methods using highly variable genes. Using a cluster-aware, weighted consensus clustering approach, we can combine the outcomes from Gini index and Fano factor-based clustering and identify both common and rare cell types. In this new version (GiniClust3), we have substantially increased the speed and reduced memory usage in order to meet the need for large data size. It can now be used to identify rare cell types from over a million cells. Previous versions of GiniClust can be found below: GiniClust https://github.com/lanjiangboston/GiniClust. GiniClust2 https://github.com/dtsoucas/GiniClust2.
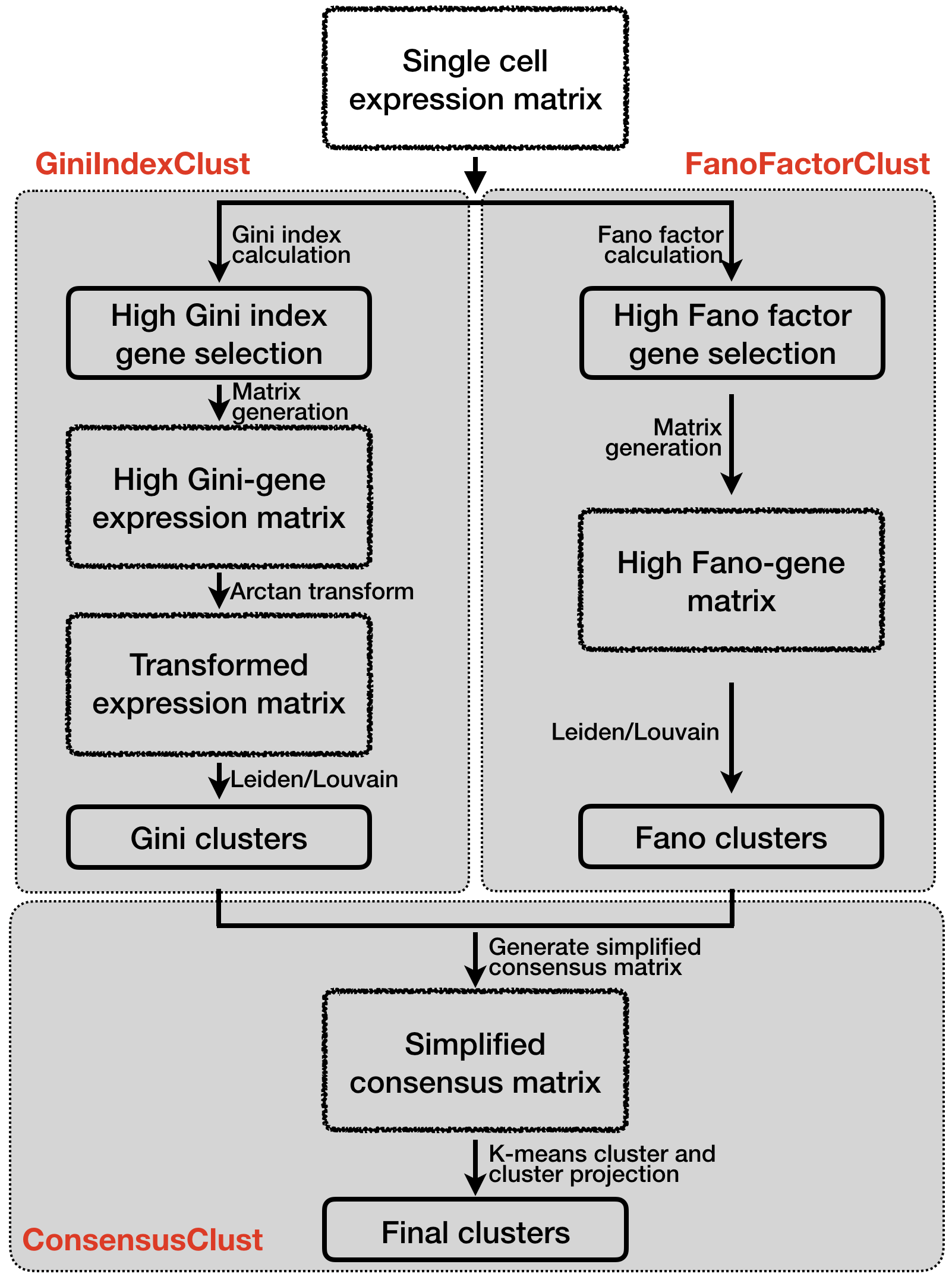
A schematic overview of the GiniClust3 pipeline
Citation
Dong R, Yuan GC. GiniClust3: a fast and memory-efficient tool for rare cell type identification[J]. bioRxiv, 2019: 788554.
Tsoucas D, Yuan G C. GiniClust2: a cluster-aware, weighted ensemble clustering method for cell-type detection[J]. Genome biology, 2018, 19(1): 58.
Jiang L, Chen H, Pinello L, et al. GiniClust: detecting rare cell types from single-cell gene expression data with Gini index[J]. Genome biology, 2016, 17(1): 144.